Dunn Research Group
The generation and detection of mechanical force is a central aspect of cell and developmental biology. Cells sense their physical surroundings by pulling on each other and the extracellular matrix (ECM). The resulting physical cues allow cells to communicate with each other, to coordinate complex collective movements, and to make critical decisions about cell growth and differentiation. Despite this central importance, the mechanisms by which cells exert and detect force remain poorly understood both in single cells and in whole organisms.
Our goal is to understand how cells generate, detect, and respond to tension at the molecular level. To do so, we are using new microscopy techniques that allow us to measure mechanical forces inside living cells, and even in whole organisms. The results from this endeavor will be highly relevant to many aspects of basic research and human health, including heart disease, cancer metastasis, and the development of stem cell therapies, all of which are governed in part by the mechanical interactions of cells with their surroundings.
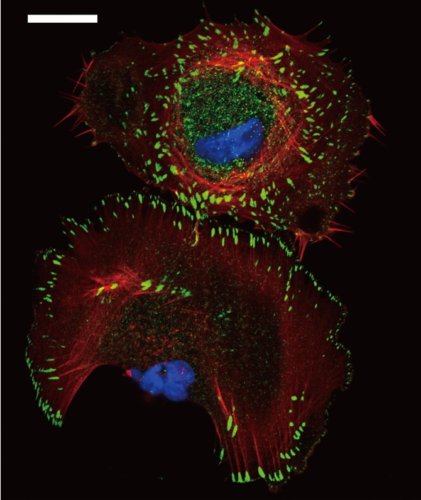
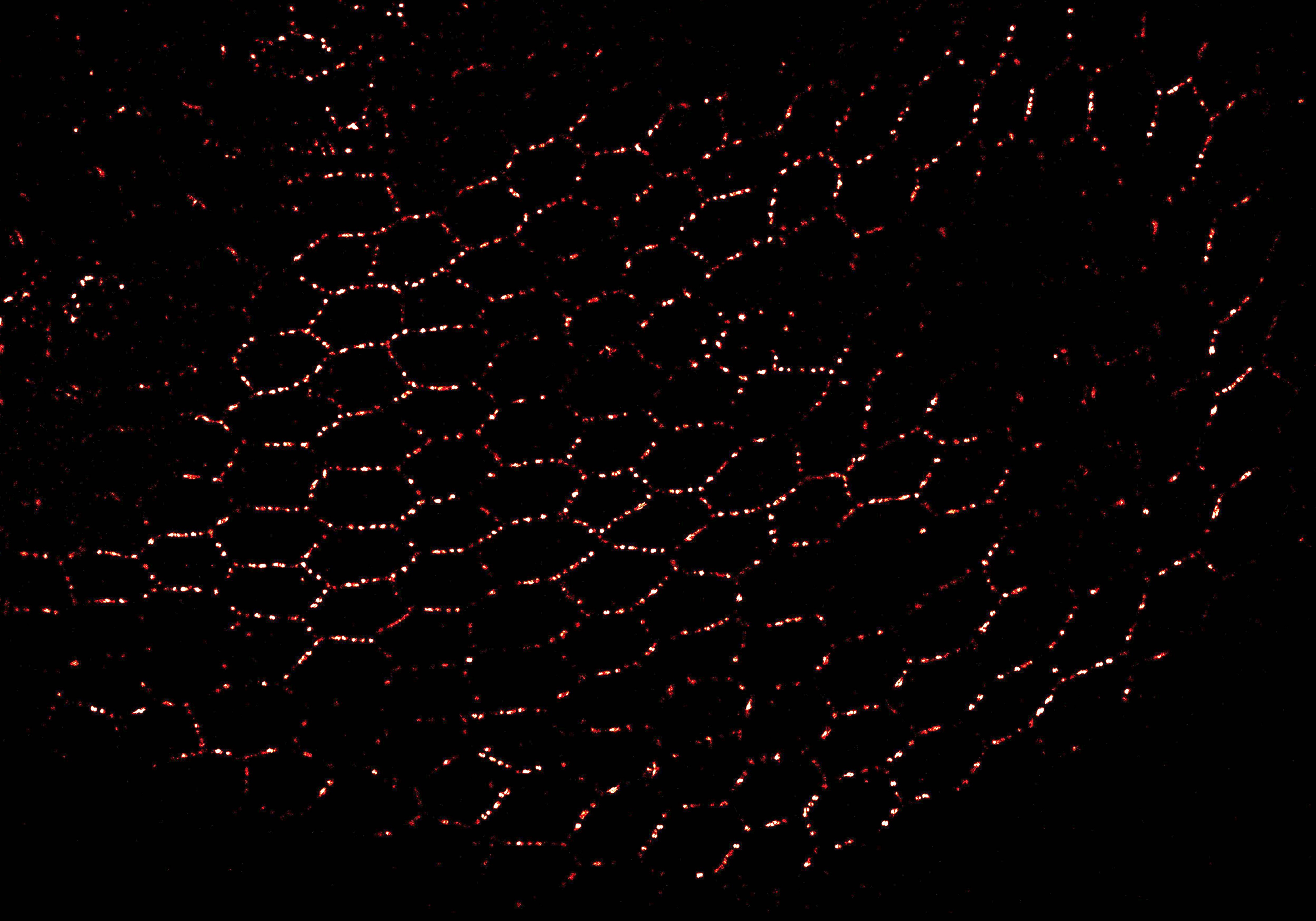
During embryonic development, many epithelial cells must be oriented relative to their surroundings. Directing the cells requires activity of the planar cell polarity (PCP) signaling pathway. This pathway provides directionality within the layer of cells and distinguishes one end of the developing tissue from the other. The molecular mechanism underlying PCP signaling remains poorly understood. Therefore, we use advanced microscopy techniques to study how molecular interactions are responsible for the PCP pathway inside living organisms. In collaboration with the the Axelrod Lab, we use fruit flies that have a suite of highly developed genetic tools and exhibit a well-characterized PCP patterning in their wings. Acquiring an increased understanding of the PCP pathway may one day contribute to preventing developmental anomalies such as heart and neural tube defects.
People in this line of research: Silas
Adhesion G-protein coupled receptors (aGPCRs) are a class of membrane receptors that are critical in several developmental and disease pathways. Despite their importance, the mechanisms of signaling activation across this diverse class of molecules are largely unknown. We are interrogating the mechanisms of aGPCR activation with biophysical and biochemical tools. We have applied forces on engineered aGPCRs with magnetic tweezers, and are currently employing live-cell imaging techniques to investigate the effects of clustering and allostery on aGPCR activation.
People in this line of research: Abrar, Chuqiao, and Brian
Cells are subjected to a wide variety of chemical and physical cues from their microenvironment that signal for shifts in gene expression to coordinate specific cellular responses. Significant focus has been placed on how biochemical signals impact fundamental cellular processes such as stem cell differentiation and cell migration, yet mechanical forces are known to impact these same processes and their roles remain poorly understood. In collaboration with the Wysocka Lab, we are utilizing various imaging techniques (such as live-cell imaging and correlative light and electron microscopy) in concert with biophysical and biochemical approaches to determine how mechanical force impacts chromatin structure and organization in both mouse and human embryonic stem cells.
People in this line of research: Emma
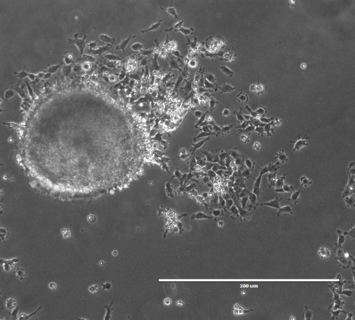
Cryogenic electron tomography (cryo-ET) is the highest resolution 3D imaging technique available to the life sciences, capable of visualizing molecular complexes in their native cellular environments at nanometer resolution. We previously developed a technique to improve the throughput of cryo-ET data acquisition at cell-cell junctions through micropatterned electron microscopy grids, and we are currently applying this technique to study heart muscle cells, or cardiomyocytes (CMs). Mutations that affect the contractility of CMs are implicated in numerous cardiomyopathies, but the basis of pathogenesis is poorly understood. In collaboration with the Dahlberg and Bernstein Labs, we are determining the ultrastructural ramifications of disease-associated mutations and investigating how such structural changes could lead to disease.
People in this line of research: Joey
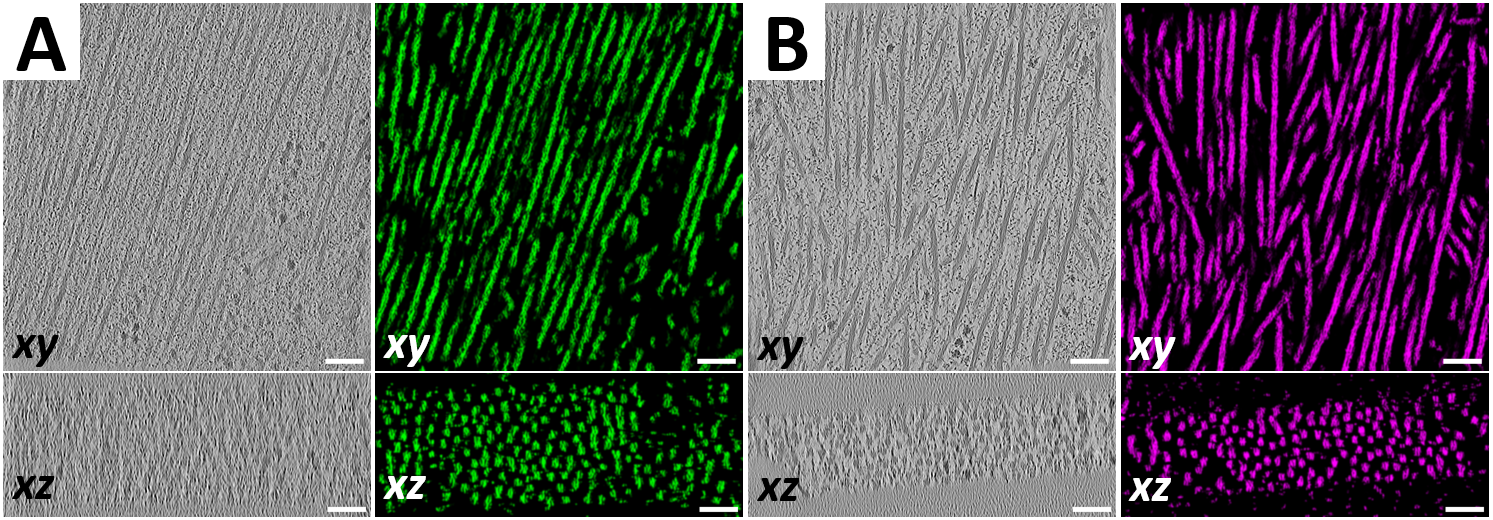
The basic contractile unit of muscle cells is the sarcomere, consisting of alternating bands of thick (myosin) filaments and thin (F-actin) filaments that slide past one another during contraction. The Z-disc is a highly ordered protein complex that links the barbed ends of F-actin in adjacent sarcomeres, allowing for the transmission of tension through sarcomeric contraction. The precise mechanism of how the Z-disc forms in the developing muscle is unknown, as is the role of Z-disc mechanosensing in development and disease. To tackle these questions, we will assess how key Z-disc proteins respond to applied forces through optical trap assays. We plan to add more proteins to the assay over time, eventually reconstituting the early stages of Z-disc maturation in vitro to better understand how mechanical forces play a role in Z-disc development and mechanosensing.
People in this line of research: Chris
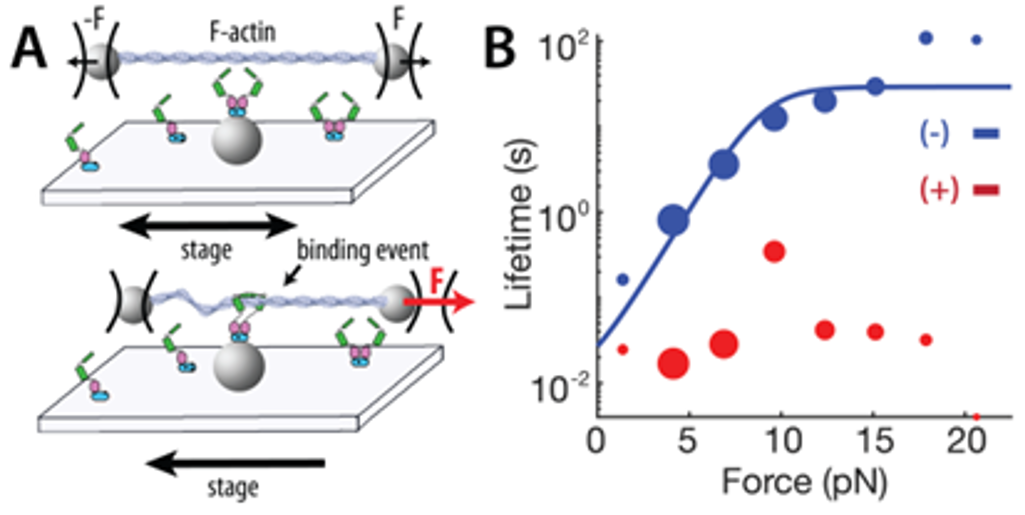
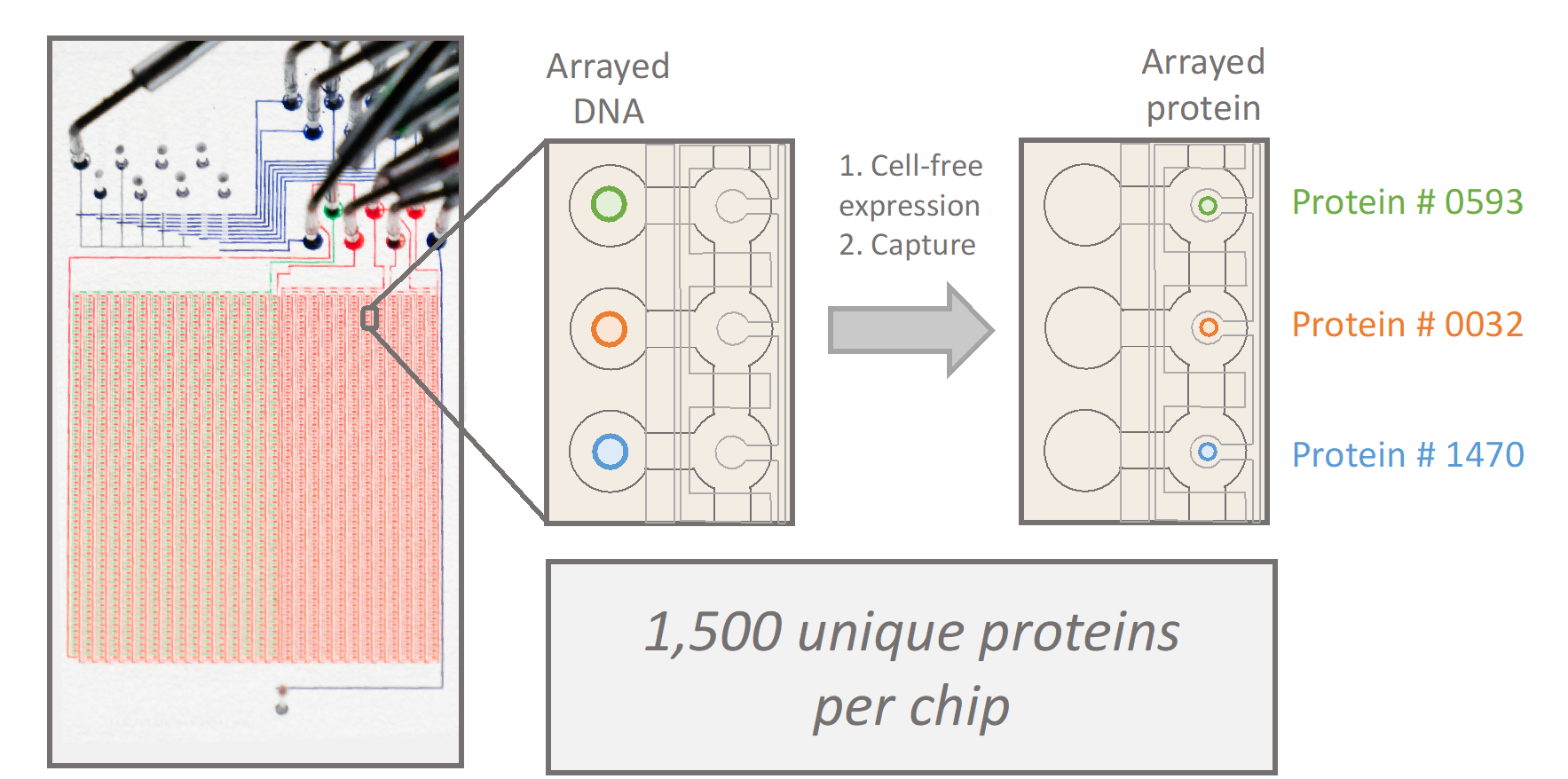
Many proteins involved in cell adhesion, motility, and contraction are sensitive to mechanical forces. In fact, a subset of proteins experience stronger interactions with their binding partners under load, exhibiting properties of a “catch bond.” Conventionally, catch bond behavior is assessed through optical tweezers and other single-molecule assays, which limits the throughput of molecular discovery. In collaboration with the Fordyce Lab, we will design and implement custom microfluidic devices to spatially array, purify, and measure 1,500 unique proteins under force. These high-throughput measurements of protein function under force will empower the discovery, engineering, and characterization of force-sensitive proteins, and mutational scanning may reveal molecular determinants of catch bonds and their biological relevance in vivo.
People in this line of research: Matt
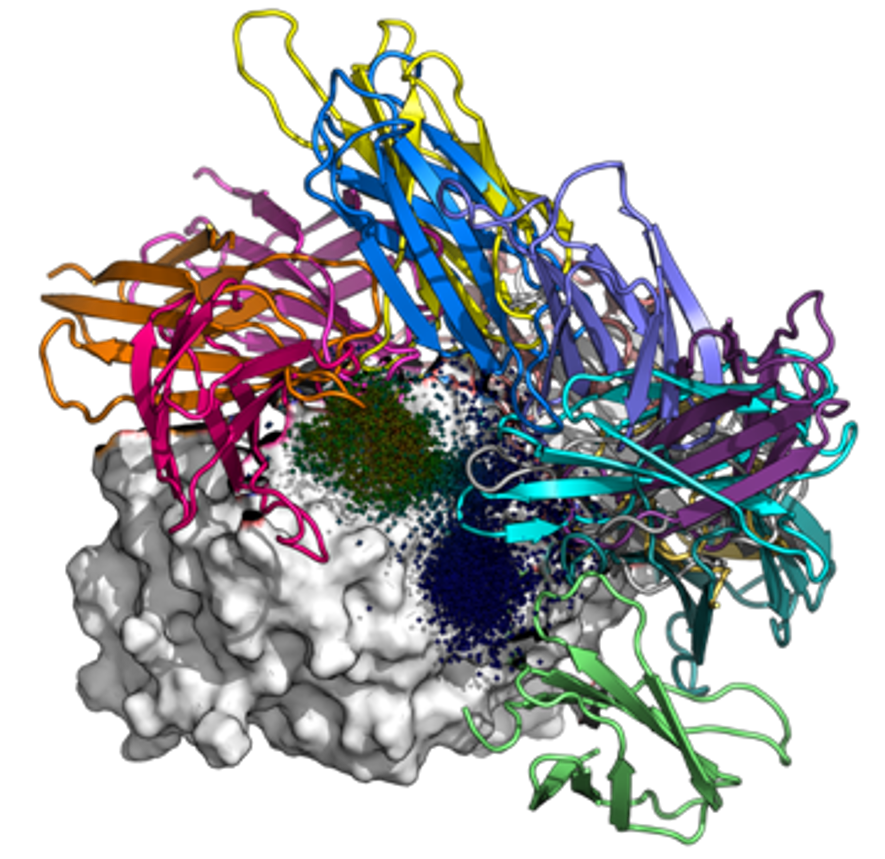
Malaria is a major global health burden; approximately 250 million contract the disease and 600,000 succumb to it each year. To fight malaria, there is a strong need for sensitive rapid diagnostics tests that can detect conserved antigens in asymptomatic cases. In collaboration with the Huang Lab, we are designing binders against parasite lactate dehydrogenase (LDH) in silico. We aim to obtain reagents with picomolar binding affinities – several orders of magnitude better than monoclonal antibodies – by creating fusions of monobodies that bind to non-overlapping epitopes.
People in this line of research: Paul
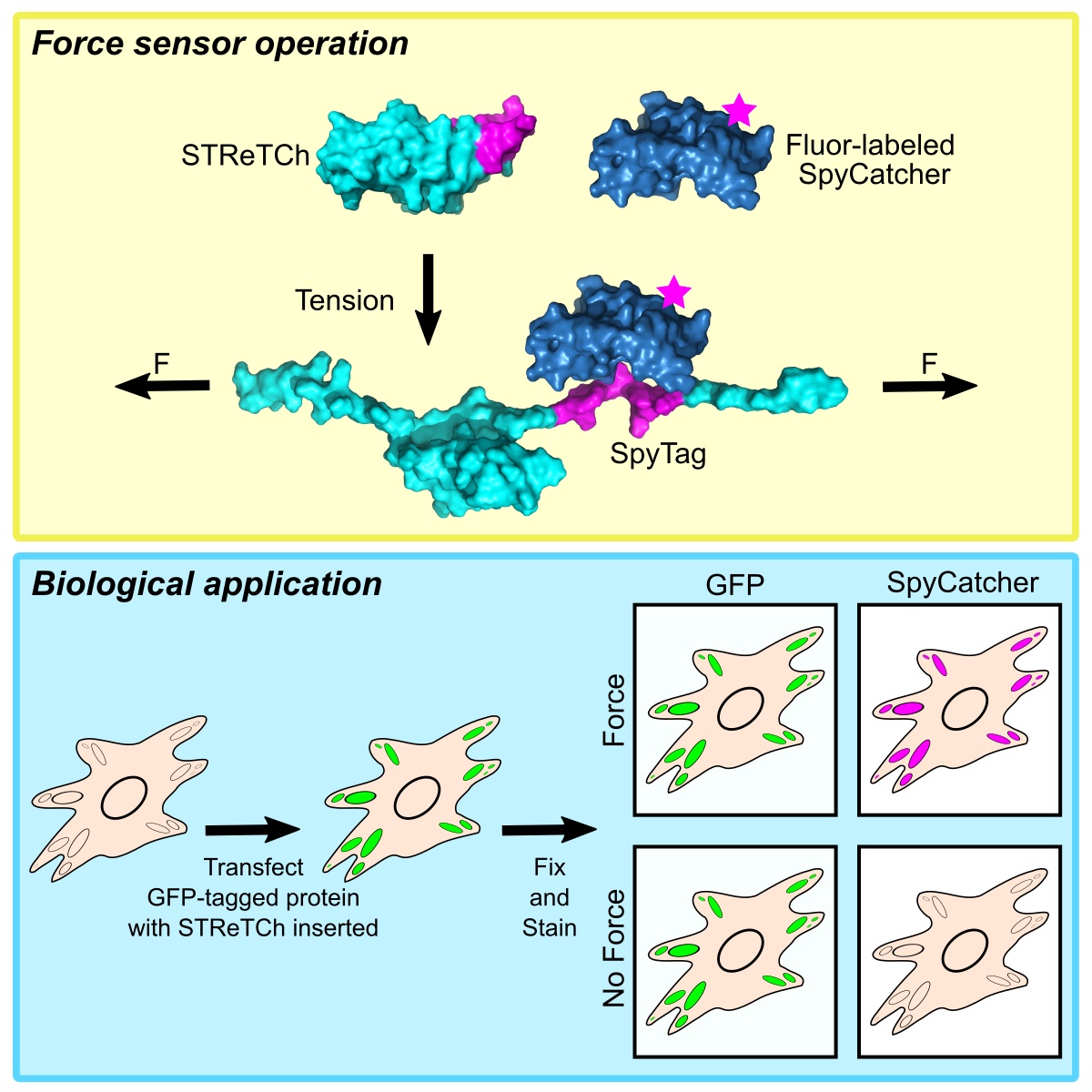
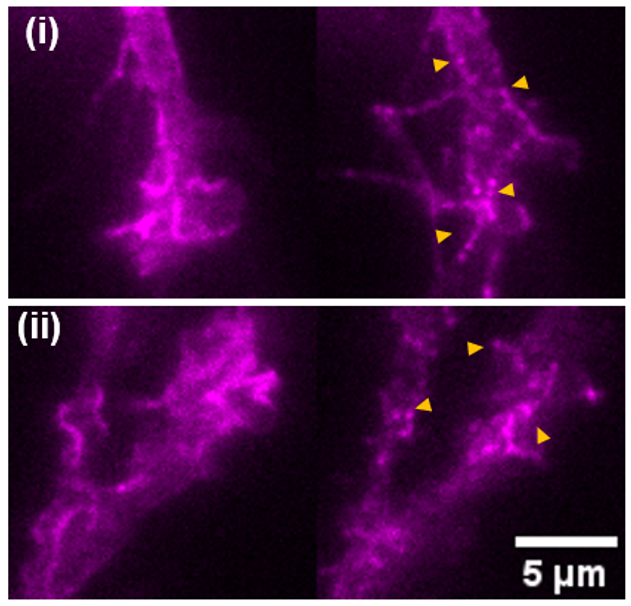
While existing FRET-based force sensors have yielded many insights into the ways by which forces are transduced across proteins, the complex optical setups and data analysis required for measuring and interpreting FRET have limited their use. We have engineered a compact, 11 kDa molecular tension sensor termed STReTCh (Sensing Tension by Reactive Tag Characterization) that does not rely on experimentally demanding FRET-based measurements and whose use follows typical fix-and-stain protocols. STReTCh reliably detects forces above 2 pN, making it one of the most most sensitive molecular force sensors described to date. As proof-of-concept, we have demonstrated that an extracellular STReTCh-based sensor reliably visualizes cell-generated forces at integrin-based adhesion complexes. In addition, we have incorporated STReTCh into vinculin, a cytoskeletal adaptor protein, as a genetically-encoded sensor of intracellular force and show that STReTCh reports on forces transmitted between the cytoskeleton and cellular adhesion complexes. We are currently building upon this work by engineering new variants of STReTCh to enable streamlined, live imaging of force in cellular systems to probe the spatiotemporal dynamics of force transmission and improve the accessibility of force measurements in biological systems.
People in line of research: Brian